Research that laid the foundations for research as diverse as understanding cancer, prion diseases, and malaria as well as offering hope of faster diagnostics and improved food quality control has been awarded this year’s Nobel Prize for Chemistry.
John Fenn of Virginia Commonwealth University, Richmond, USA, and Koichi Tanaka of the Shimadzu Corporation, Kyoto, Japan, share half the 10m Swedish Krona prize for their work on methods of suspending proteins and other macromolecules, such as polymers and nucleic acids, for analysis by mass spectrometry.

John Fenn
Mass spectrometry can be used to determine the mass of a molecule or indeed fragments of that molecule. Fenn began work on devising a method for suspending proteins in a mass spectrometer in the 1980s. Until that time mass spectrometry was only really useful for small molecules with a limited number of atoms not the vast macromolecules of nature, such as proteins, which can have hundreds and thousands of atoms.
Fenn’s method of electrospray ionisation (ESI) was based on spraying droplets of protein solution with an electric field and allowing the solvent in which they were dissolved to evaporate. This process left stark, naked protein ions hanging in the chamber of his mass spectrometer ready for analysis.

Koichi Tanaka
Tanaka, meanwhile, had discovered that a pulse of laser light could blast a viscous sample of protein into separate charged molecules. Once in this form, he used an electric field to accelerate the ions across the mass spectrometer’s chamber. He could then build up a spectrum of the original protein molecule based on the differences in the times of flight recorded for each electrically charged fragment.
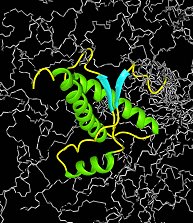
Protein structure
Many sensitive mass spectrometry techniques, such as MALDI (matrix-assisted laser desorption ionisation), emerged following this pioneering work.

Kurt Wüthrich
While Fenn and Tanaka provided scientists with a way to find out what protein they had in a sample and how much, their work said nothing of what that protein looks like – its three-dimensional molecular structure, in other words. That was down to the work of the winner of the other half of this year’s chemistry Nobel – Kurt Wüthrich of the Swiss Federal Institute of Technology, Zürich), Switzerland.

Kurt Wüthrich talks about the protein structure
Wüthrich developed a way to assign each signal in a proton nuclear magnetic resonance spectrum to the appropriate hydrogen atoms – and there are many of them – in a protein. A comparison of the distances between pairs of peaks in the spectrum could then be used to build a picture of where they all fit in the structure. This information can then be used to generate the protein’s all-important three-dimensional structure.
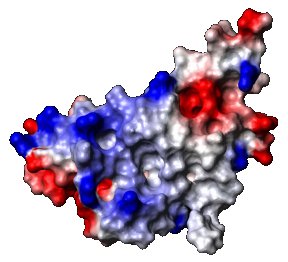
Another view of the protein
The technique works on proteins in solution so, unlike X-ray crystallography, does not require a crystalline form to be made first. The method even solves proteins working in living cells. Since the invention of the method in 1985 hundreds and hundreds of the thousands of known protein structures have been determined by NMR.
Further reading
John Fenn
http://www.has.vcu.edu/che/people/bio/fenn.html
Shimadzu Corporation
http://www.shimadzu.com/
Kurt Wüthrich
http://www.mol.biol.ethz.ch/wuthrich/people/kw/
Suggested searches
Nobel prizes
Electrospray ionisation
Proton NMR spectroscopy
Protein structure